Thinking about the future of an increasingly biologized world, we see ourselves at the intersection of the scientific and algorithmic worlds. We aspire to galvanize and connect other folks at this interface so are building a hands-on collective which is focused on building the next generation of companies spanning these worlds (more here; join here!). This program kicks off with a breakfast on April 21st in NYC, and includes access to a tailored Discord community, quarterly 1-on-1 sessions to understand where we can help (e.g. directly connecting you to co-founders to start a company). We're also planning to have collective-wide quarterly calls where you can meet other ambitious folks and trade ideas. Connecting the dots between tech and bio
With this blog post, we’re delineating the progress of generative machine learning models (diffusion models, transformers, LLMs, and multimodal algorithms) applied to bio. The shift towards using these models on biological training data has enabled biological breakthroughs from understanding the syntax of proteins to generation of entirely new proteins. We are still limited, however, by the relatively small amounts of training data in bio and the current limits in experimental throughput and noise. Despite today’s limitations, we should still be imagining where these models, trained on copious amounts of quality biological data, could take us. We believe that the transfer of learning from algorithms in tech to bio won’t always be the direction of innovation. With cheaper experiments and growing utility of bio (both for climate and health), we think that we’re on the cusp of the fundamental algorithms being used on bio data first and then being translated into pure tech. This is one of the many reasons why we’re developing this community (learn more here; apply here).
Models from tech in bio
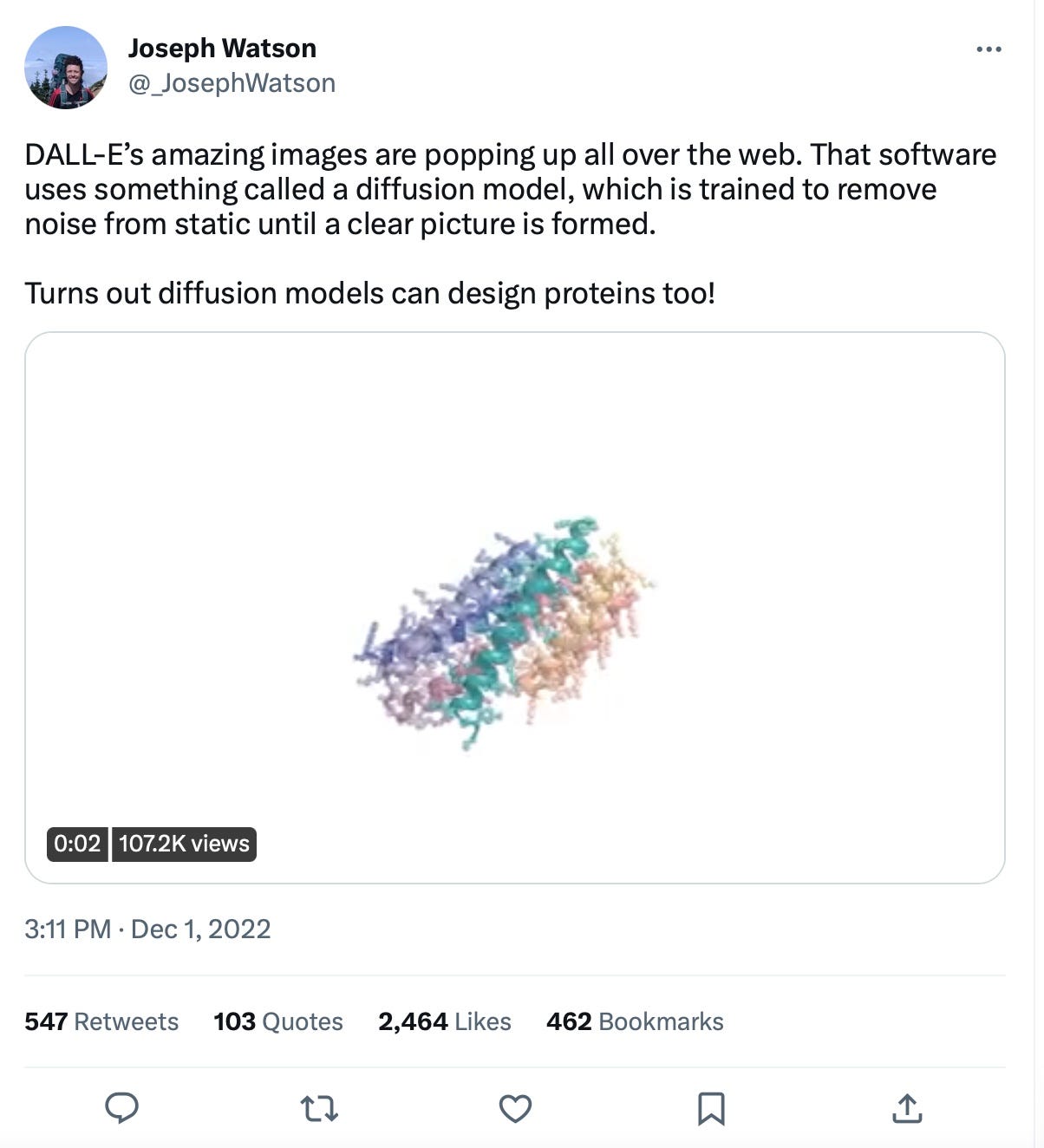
Diffusion models made waves in the tech world with the text to image generation (Midjourney, DALL-E 2, Runway) and with music generation (Magenta Project). The same process of diffusion (e.g. noising and denoising to data to generate new objects) has been used in biology with RF Diffusion, hacked AlphaFold (instead of the Rosetta Fold of RF Diffusion) and diffusion combinations (hack here) and sequential diffusion (here). We’re imagining how quickly we can go from combined structure and diffusion based prediction of single proteins to metabolic pathways - e.g. generative metabolism. This could come from multimodal data which encompasses the protein function + search optimization for protein families which allow this (or something similar) reaction. Someday we could even expand the search space to enable the conversion of completely new molecules using protein catalysis. In the words of Stephan Heijl:
‘Just enter a resource and target molecule, let the servers run for a bit and test the resulting pathway to find your required complex molecule being made in the lab!’There are iterations of diffusion models that are also making in-roads in DNA (see DNA diffusion here). This work is spearheaded by OpenBioML and is working towards understanding what regulates DNA expression (which governs which mRNA and consequently proteins are expressed.) This is relatively overlooked compared to protein folding but is absolutely essential for the full realization of synthetic biology through regulation of how much valuable product is made.
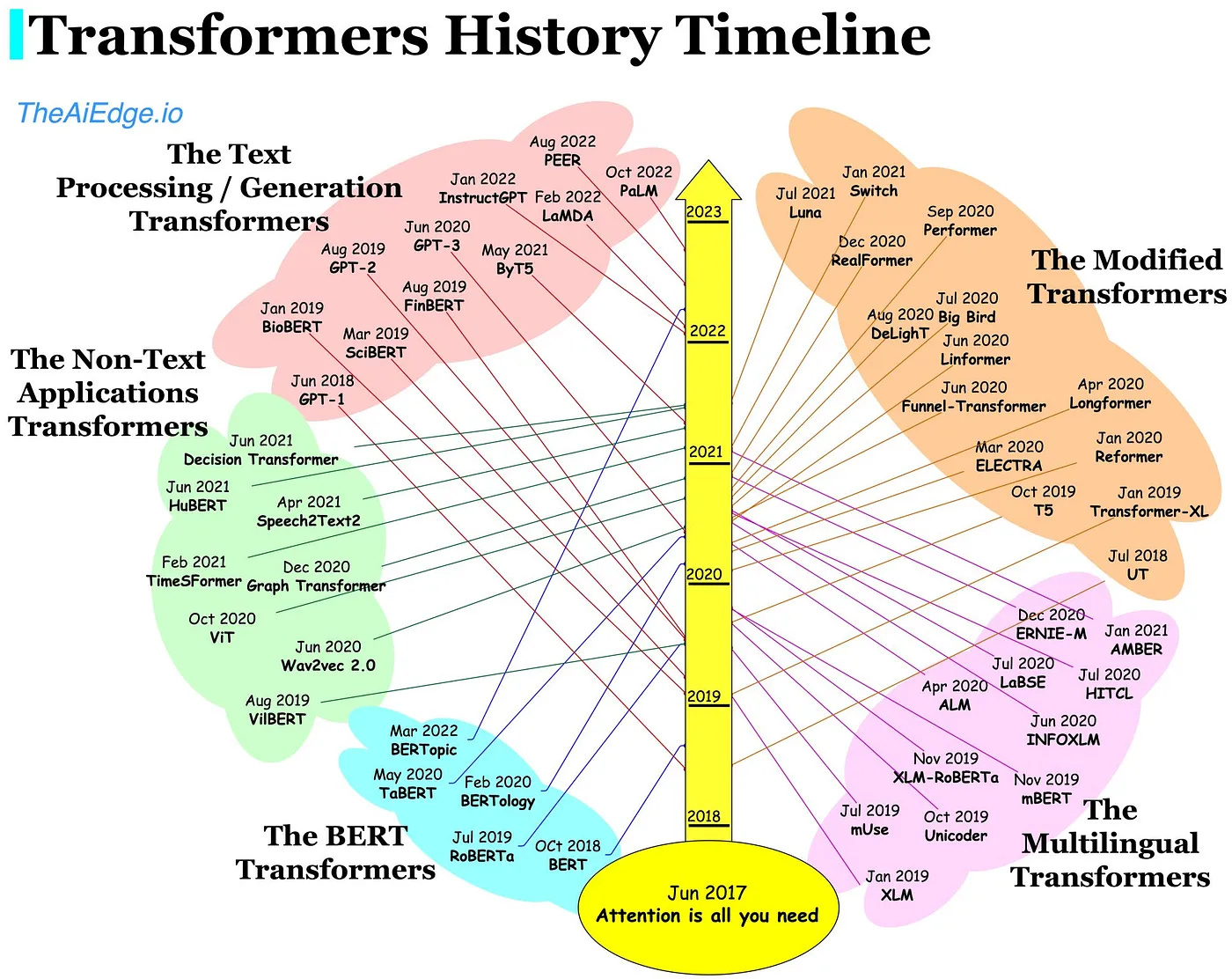
At the same time Large Language Models (LLMs) have made a splash with their uncanny ability to create and synthesize text. Meta’s leaked LLaMA, GPT-4, Gopher, amongst others have continued to outperform expectations, writing compelling poetry, and books even in lightning time. LLMs are enabled by transformers which allow for the parallelization of input sequences. Self-attention during this process facilitates focus on different parts of the input sequence creating longer-lasting ‘memories’ than in algorithms previously described. This allows for better understanding of long sequences (think sentences or proteins). These LLMs have been tailored to certain industries, like bio (meet BioGPT), and even reached human parity in ingesting biomedical research to answer questions. LLMs have even uncovered protein syntax (like ProteinMPNN) and are built upon the famous transformer paper, ‘Attention is all you need’. To continue the theme of big tech in bio, Salesforce and UCSF used language models to make functional proteins across diverse families.
Rise of multimodal data in bio
Just like AlphaFold or RosettaFold diffusion models create better protein structure predictions, we think that multimodal data in bio will continue to enable better understanding and generation of biological systems. One model to learn them all, showed that various multimodal data (think audio, text, images) could help models generalize and get close to state of the art performance in many different tasks, using just one model. Since this publication multimodal has been applied in several circumstances in bio (state of the art protein prediction, cell-type-specific functional gene networks, integration of multimodal single-cell data, and more).
Future proofing
Generative models have the potential to create many de novo proteins, DNA regulatory sequences, and molecules. To ensure these work and positively train models to reinforce the learnings, there continues to be a need for high-throughput experimentation, for both validation and learning. Close collaboration between dry and lab components, with both parts actively involved in the design of experiments, generation of datasets and evaluation of model performances is going to be key to lowering the barriers for the application of these models in the biotech industry. It is important to note that faster generation of candidates does not necessarily correlate with better, faster, or more efficient results. Therefore, there is still a need for better ways to identify candidates to advance in the pipeline, while also future proofing the business.
Potential focus areas
Whilst it’s easy to think about limitations of the current tech, we think there’s tremendous value in ideating around the trajectory of generative models. We aren’t alone in thinking that SciFi isn’t pioneering enough:
And love the idea of generative butterflies:
In that vein, we’ve put together a list of ambitious ideas such as:
New microbial communities could be designed to enable your gut to intake a perfectly balanced diet or provide regenerative vitamin sources
Create balanced ecosystems that could reforest deserts created from climate change, all from newly generated organisms
We could even see the first batch of organisms specifically designed to be grown on extraterrestrial planets
Prediction of unseen bacteria and microbes could be beneficial for sustainable energy solutions, following the first examples of energy produced from living organisms
Generative agents trained on cellular circuits could predict new combinations of perturbations that lead to desired outputs (e.g. production of desired substances) opening the door to the use of the same cultures for different purposes and enhancing the resilience of supply chains
Here’s our database with more ideas on where we think the future is heading.
Moats
Besides these utopian futures, we think generative models in bio uniquely provide moats that you don’t see in the tech sector. In the technology sector, the value capture for generative AI is still being determined (as discussed by a16z here). In bio, progressing molecules and proteins through regulatory milestones provides clear moats. Even in synthetic biology/industrial biotechnology which has historically been a B2B race to the bottom, companies such as Geltor, Checkerspot, and Debut Bio have also shown that owning the full value chain is possible. We’re tremendously excited for the first generative consumer products that’ll emerge from this field. In short, long-term value capture starts today with no shortcuts or work around the utility of gathering unique biological data (and lots of it!) and progression of leads. This will feed into better drugs, materials, and ultimately better ecosystems for our world and others. Because of the multi-factorial impact of these models, we believe community building between and inside tech and bio has never been more important. We’re excited to meet ambitious and imaginative founders and future founders here!
Many thanks to co-author, Fabio Boniolo! Fabio is a researcher with us at Compound and a post-doctoral researcher at the Broad Institute working on bioinformatics for childhood brain cancer. Fabio previously worked on high throughput screening methodology on primary cancer cell lines and also on digital twins for bioreactors. Learn more about Fabio here!
The problem spaces above are just a sample with all we think we can accomplish by merging the tech and bio worlds. Apply to join our ambitious collective of ideators and futurists to discuss. FAQs for the community can be found here. This program kicks off with a breakfast on April 21st in NYC, and includes access to a tailored Discord community, quarterly 1-on-1 sessions to understand where we can help (e.g. directly connecting you to co-founders to start a company). We're also planning to have community-wide quarterly calls where you can meet other ambitious folks and trade ideas.