Bio foundation models - definitions, nuance, and tactics
Transposing tech to bio and back again
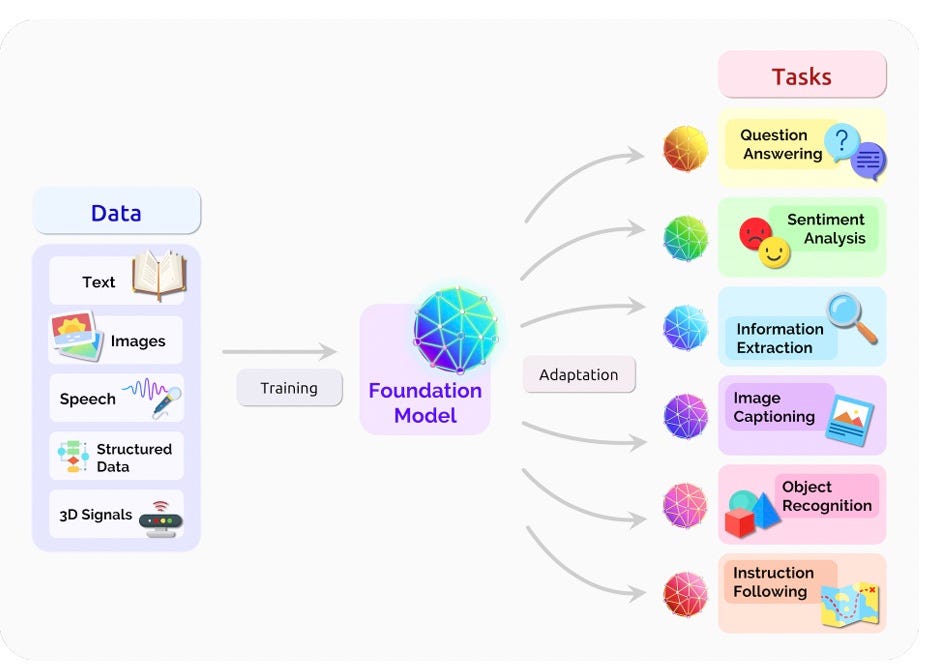
Seemingly every day we see a new publication or company in bio that’s claiming it’s the foundation model for glioblastoma, T-cells, chemistry, and others. While this is exciting, we can’t help but grapple with the nebulous terminology of foundation models, especially in the context of bio. In this blog we talk about what foundation models are defined as in the context of tech (training data size, parameters, sociological context etc) and translate this to bio. We also talk about why the commercialization of foundation models is similarly distinct from tech, centering around product instead of APIs. We believe the product focus will ultimately limit how big any company could become in this space. Finally, we highlight a database we published recently highlighting 45+ foundation models in bio.
Foundation models in tech
The term foundation models was defined in 2021 by Bommasani et al. in a seminal paper ‘On the Opportunities and Risks of Foundation Models’. I highly recommend anyone interested in the space to check out the article that argues transfer learning, computational scale, and sociology shifts are the crux of foundation models. Scale makes foundation models powerful through hardware improvements (GPU throughput and memory advantages), transformer model architecture and materially more training data. However, previous terms such as pre-trained and self-supervised model didn’t encapsulate the ‘sociological impact’, conferring a shift in AI research and deployment. (The sociological impact being the shift of self-supervised learning from a ‘subarea’ to a ‘substrate’ - where one model could be useful for wide ranging tasks.) The foundation models they mention range from purely natural language process (NLP) models such as BERT to multimodal datasets as are standard in healthcare (medical images, clinical text, and structured data). For the authors, BERT (Devlin et al. 2019) was the watershed model which became the substrate for the field of NLP. I think it’s important to highlight the scale of BERT as a reference point for other models being called ‘foundation models’.
The base BERT of Devlin et al. 2019 had:
110M parameters
12 layers/transformer blocks
768 hidden size
12 self-attention heads
Pre-training data: 3BN words from Book Corpus and English Wikipedia.
The large BERT of Devlin et al. 2019 had:
340M parameters
24 layers/transformer blocks
1024 hidden size
16 self-attention heads
In my mind, these technical specifications give a technical grounding of the size of foundation models but think the sociological element is crucial and more context specific in definitions. Namely, if the model is as large as the parameters above but not able to be used for a variety of downstream tasks, I don’t think that’s a foundation model.
Graphic from Bommasani et al. 2021.
Foundation models in bio
There have been several ‘foundation’ models in bio and healthcare including AlphaFold (protein structure prediction), RoboRx (predicts drug prescription interactions), DeepNovo (peptide sequencing from mass spectrometry), EcoRNN (ecosystem population dynamics), amongst others. As with canonical foundation models (BERT, GPT-3, PaLM, LLaMA, LaMDA, Orca, Claude, and others), the bio foundation models provide a base layer which can be fine-tuned for specific purposes. For example, AlphaFold has been fine-tuned on specific protein families such as MHC Class I and Class II and T-cell receptor:MHC complexes, amongst others to help increase binding specificity or model accuracy. The models above are mixed between single and multimodal but see a trend line towards multimodal bio models becoming the norm and where we envisage emergent properties being observed. Given that bio is inherently multifaceted and complex, it makes sense that models incorporate more modalities especially as AI will perhaps be able to find correlations and factors across modalities that humans simply do not have the ability to at scale. This approach is corroborated by a recent Forbes article about the ex-Meta ESM team claiming they would build a model to integrate other biological data from DNA sequences, gene expression and epigenetic states. Moreover, the tailwinds of healthcare foundation models (Med-PaLM, CLaM, FEMR, ClinicalBERT, ehrBERT, bioGPT, and others) give credence to the multimodal trendline in bio too. (An interesting part of the healthcare foundation models is that they’re made by forking base models such ast BERT, PaLM, and GPT.)
Clinical and biomedical foundation models fro Winow et al. 2023.
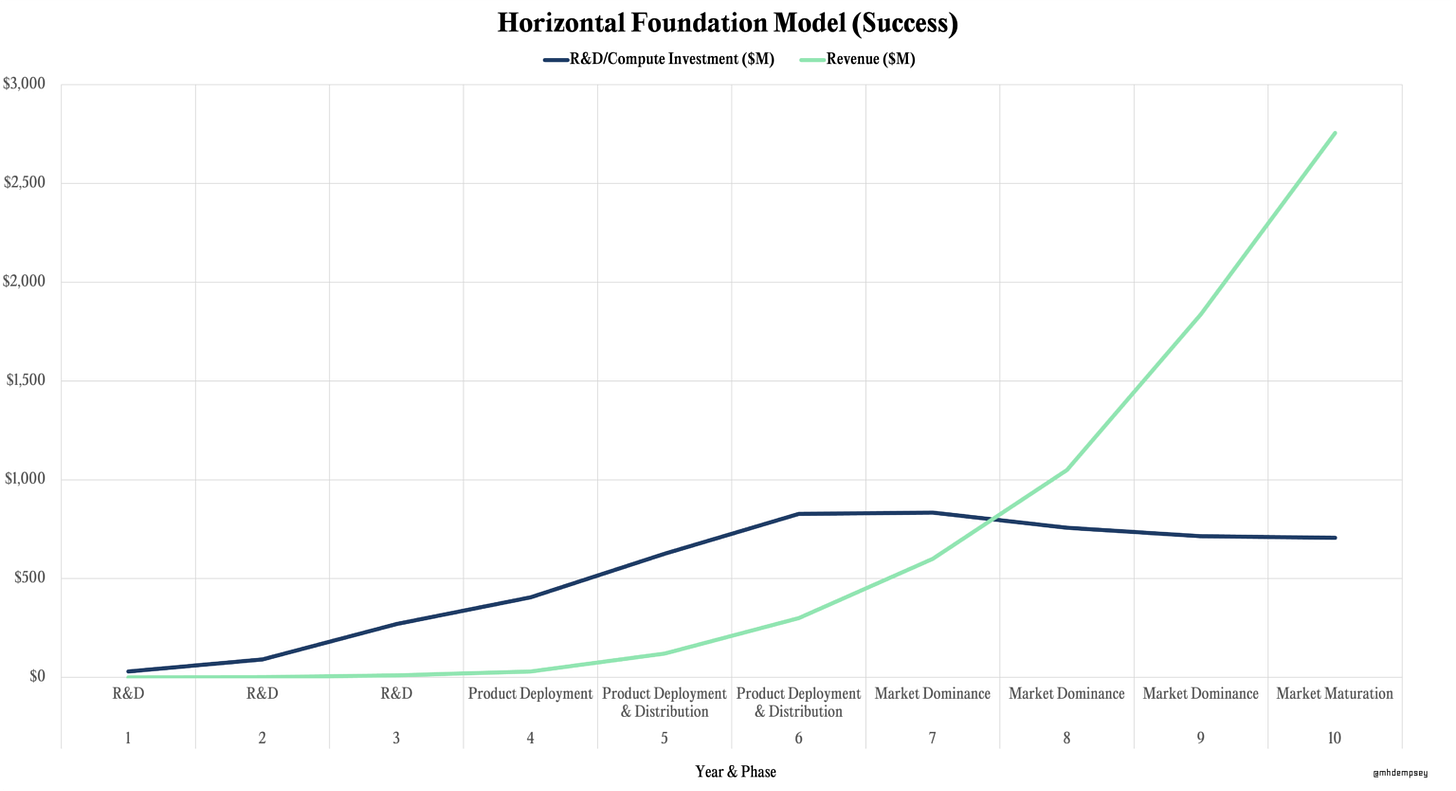
Despite the tantalizing utility of multimodal data in bio, it’s important to highlight some single-modal areas of research. Something that’s been a surprise with protein folding is the *shockingly* small amounts of data to be needed to train some models. For example, OpenFold (a trainable open-source implementation of AlphaFold2) had essentially the same performance on a training set of 10,000 sequences as it did on a training set of 100,000 protein sequences (which was the entire protein databank). Even a training set of 1,000 sequences, led to a structure prediction fidelity of 76% of the model trained with 100,000 sequences. This is incredible because it means the tacit laws of protein folding are seemingly more easy to learn than the laws of human language. Tactically, it means much less data could be required for foundation models in bio than are needed in tech which could save significant money and time in compute and data generation for some biological modalities. In a recent blog post my colleague Mike Dempsey, delineated the cost discrepancies of horizontal foundation model (billions to tens of billions cumulatively and 6+ years) versus vertical foundation models (hundreds of millions cumulatively), which speaks to some of the profitability and R&D dynamics of more vertically focused foundation models (see his graphics below).
In the case of more vertical foundation models (such as single modal biology models), Mike shows the much lower capital requirements (~$100M) needed to get to profitability and market dominance. Despite the cost savings possible by using smaller datasets, increasing the performance and data moat is at the heart of many companies. Companies such as Gandeeva Therapeutics and Generate Biomedicines are heavily investing in cryo-electron microscopy (CryoEM) hardware which experimentally determines protein structures, which adds proprietary data to their models. CryoEM is a very strategic decision of these companies as well given that it works best for large protein complexes (which are notoriously difficult for models), allowing the protein movement (conformational states) to be captured, all in native environments since it’s based on snap freezing the protein (as opposed to perturbed environments such as those required to make protein crystals). The bar for many other companies replicating this strategy is difficult given the price of CryoEM machines ($7M) and cost to keep them running ($10,000 per day). This circles back to another internal thesis we have on full-stack deep tech businesses which we believe ‘enable them to be significantly larger than the largest incumbent’.
Outside of protein structure, an area of tremendous interest has been single-cell RNA sequencing (scRNA-seq). There’s increasing size of available datasets (e.g. CELLxGENE or the Human Cell Atlas) spanning millions of cells from multiple tissues and disease states and their potential to create values throughout the drug development pipeline. Multiple foundational models have been introduced in the space, often implemented after adapting training strategies and architectures that have been successful in NLP to the peculiarities of biological data, such as sparsity and the continuous nature of molecular measurements. These applications have shown the potential scRNA-seq based foundation models have to tackle a variety of routine tasks such as cell-type annotation, batch integration, perturbation prediction, and drug response prediction. In the list of foundation models below we see the breadth of architectures (encoder- or decode-only to full-stack encoder-decoder transformers) and training strategies implemented, similar to the variety observed in the most common large language models. In general, these models are pre-trained on at least 5M cells in a self-supervised fashion and further fine tuned for downstream applications, and share some of the properties of LLMs, e.g., scaling law. Gene expression profiles are treated as “sentences” where each gene is modeled as a token. We highlight the different choices taken to transform expression values to discrete tokens that can be further embedded and used for training. We envision this step, together with the identification of the most meaningful architecture and masking strategies for biological applications, as critical in future iterations of foundation models in tech bio, as a thorough evaluation of these approaches will be important to maximize value extraction from the training datasets and the model themselves.
Another current limitation of the field is the difficulty in directly comparing size and scale of biology foundation models since the parameters, layers, and attention heads aren’t always reported. However, in our initial compiling of the literature, there seems to be the 100M parameter size minimum for foundation models with 5M cells minimum training size for scRNA-seq data. (Although with proteins the smallest training set is hard to quantify given the great performance of OpenFold being trained on 1,000 protein structures.) Large strides are being made seemingly every day with models such as xTrimoPGLM showcasing 100BN parameter models with 1TN training tokens. We feel that the term is many times still used loosely and hope these numbers and context on the sociological element of foundation models will help folks see through the noise.
Monetization and business models
We’ve seen early signs of commercialization, companies both offering foundation models as a service for specific companies and those building companies around their proprietary foundation model. We’ve said before that bio is different from tech in terms of its moats, with biology having much more defensibility given the need for patents and regulatory milestones. We continue to believe this and see two bifurcating business models in biology - API and product. Foundation models in tech are primarily commercialized through API plugins. While a company could be built here, we think, in the first instance, the pool of potential customers is too small to support an API business in bio. We think maximum value accrual is centered around companies that are building products (therapeutic peptides, small molecules, enzymes, and materials) around their models. There might be a case for domain specific talent to fine-tune existing models for their own use-cases (TCR:MHC specificity) which could be big successes in themselves. Still, this uniquely centers around productizing which is much harder because you need talent making the foundation models (bits) and deploying the findings in the real world (atoms). The real world deployment means that bio companies will be ultimately capped (Eli Lilly - $498 BN market cap; Merck - $269 BN market cap; Pfizer - $200 BN market cap; Amgen $140 BN market cap) as compared to tech companies (Apple - $2.79 TN; Alphabet - $1.69 TN). Despite the potential size of a company being capped by manufacturing, regulatory milestones, and sales teams we think this is where a generational company, with defensible moats is made. After a large product-focused bio company is built, there could be added API-type functionality which allows for broader tech distribution.
To get to this ultimate conclusion of product-centered companies for foundation models, we first transposed the ‘tech’ versions of foundation models to bio. The sociological context requirement of foundation models is not to be overlooked in bio but still takes a narrower form (e.g. AlphaFold is always used to understand protein structures but can be fine tuned to better predict structures of certain protein classes). For this to have the highest permeation possible, the company would need to be premised upon a multi-modal foundation model, thus applicable to many downstream use-cases. If you’re building new models, infra, or companies in this space we’d love to hear from you!
Database
As we’ve been thinking about this space, we’ve been inspired by those tracking the new models (multimodal tech models, healthcare foundation models here).
We’ve launched a list tracking the new foundation models for bio. Let us know what we can add here!
Thanks to Mike Dempsey for your help in editing this piece.







What pathways to revenue do you think are exciting or overlooked for foundational models / tailored models trained on biomedical text?